Rare diseases are rarely studied in-depth, and rare cancers even less. Tackling this deficit of information, and understanding one rare, aggressive cancer has become the focus of research for Dr. Reety Arora at the National Centre for Biological Sciences. With collaborators at the ACTREC–Tata Memorial Centre and the University of Michigan, Dr. Arora recently analyzed 5 MCC cases for specific integration sites of their causative virus and delved into the data of 123 cases of Merkel Cell Cancer.
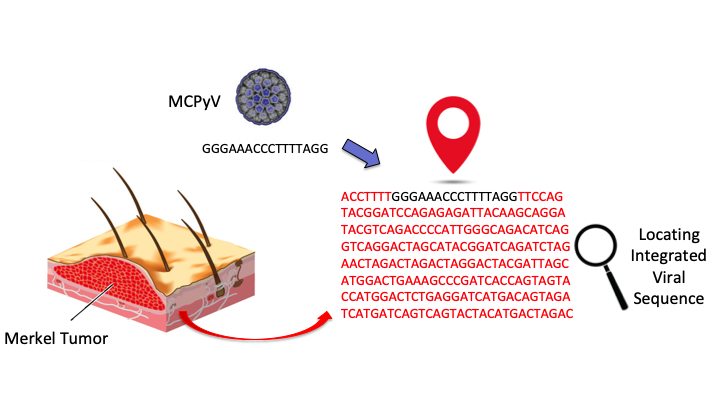
Merkel Cell Cancer is a lethal cancer of the skin, caused by either UV-linked mutations or the Merkel cell polyomavirus (MCPyV). The MCPyV was discovered in 2008 and we are still learning about the virus. Though it is commonly found, it is usually not harmful to us, and causes cancer only after integration into the human genome, followed by mutations of itself. Since the virus is found ubiquitously on human skin, detecting its presence in tumours could often be unrelated. Detecting the integration of the virus DNA into the genome is a better clinical indication.
Following a first-of-its-kind study of the cases of MCC in India, and efforts to test for and identify the MCPyV in patients as the cause of the skin tumours, the recent publication in the journal Viruses builds significantly on our limited knowledge of this cancer-causing virus. In computational genomics, large and high-quality data sets are important. The disease being rare and with limited high-quality tumor samples, there are only a few studies worldwide analyzing the integration of the virus DNA into the human genome. Also, the identification of MCPyV integration in Merkel tumors is technically challenging as the circular genome of the virus does not linearize in a consistent manner.
To carry this out, the researchers modified a bioinformatics program, created by Dr. Chandrani (one of the authors) which was used previously to detect the Human Papilloma Virus (HPV) in the human genome. They used whole-exome MCC datasets generated at the University of Michigan by Dr. Paul Harms’s group and were able to predict and validate MCPyV integration sites for 5 cases. The validation of the integration site was done via PCR testing of DNA from the original clinical samples. The researchers also collated data from a total of 123 previously published MCC cases to identify locations of Merkel cell polyomavirus in the human chromosomes. The code for the program is available for everyone to access on GitHub.
Dr. Arora says, “Integration in virally caused cancers increases the risk of cancer beyond simple viral infection as it can disrupt and deregulate nearby human cancer-associated genes, create fusion mRNA and proteins, and contribute to genome instability. While there was no definite pattern to the locations of MCPyV integration on the human genome, our study definitely showed some very interesting commonalities." On the Human Genome, the most frequent breakpoint was on chromosome number 5 (21 occurrences) followed by Chromosome 6 and 1. While there were several other integration sites on the genome, no breakpoints were found on the 21, 22, and X chromosomes. On the viral genome, the point where the DNA broke the most number of times was the part that coded for the Large T Antigen.
Dr. Pratik Chandrani, co-lead of the study, says, "The computational analysis showed that in one sample, the MCPyV DNA had inserted itself into the part of the human genome that codes for the KMT2D gene. This is the first report of involvement of KMT2D in MCPyV integration in MCC, as it has generally been reported as frequently mutated in MCPyV negative Merkel tumors." The KMT2D Gene is a tumor suppressor gene. If that gene is inactivated the chances of tumours developing increase greatly. This also means that there is an increased cancer risk if this gene is inactivated. Although this was only one observation from a single sample, this is definitely something that is worth looking into as more data emerges from future studies.
Dr. Arora adds, “Our analysis adds to the repertoire of MCPyV integration junction data and will hopefully advance the understanding of virus-mediated tumorigenesis and reveal therapeutic vulnerabilities. We are in the process of expanding our dataset for this rare cancer and hopefully can apply our analysis to genome sequencing data from Indian Merkel tumor cases soon.”
Image credit: Reety Arora









0 Comments