A new study reveals differences in the genomic history of tiger subspecies, pointing to the importance of understanding evolutionary history for future conservation
Genetic variation is like money in the bank: the more you have, the better your chances of survival in the future. Population bottlenecks decrease genetic variation, especially in endangered species. An individual's genome comprises the events that have impacted genetic variation over time, and relatively recent sequencing technologies allow us to read and interpret genetic variation across the genome. Although tigers have received significant conservation attention, little is known about their evolutionary history and genomic variation. This is especially true for Indian tigers, and with 70% of the world’s tigers living in India, such understanding is critical to tiger conservation.
A team of researchers from the National Centre for Biological Sciences (NCBS), Stanford University, and zoological parks and NGOs across the world recently completed a three-year project to gain insights into genomic variation in tigers and the processes that have sculpted it. The work, just published in Molecular Biology and Evolution reiterates that tiger subspecies are genetically distinct and reveals that although Indian tigers have the highest total genetic variation, some individuals are inbred there. Simulations based on the genomic data suggest relatively recent divergences between subspecies, and intense population bottlenecks. Analyses also indicate adaptation to cold environments in Russian far east tigers, and potential selection on body size in Sumatran tigers.
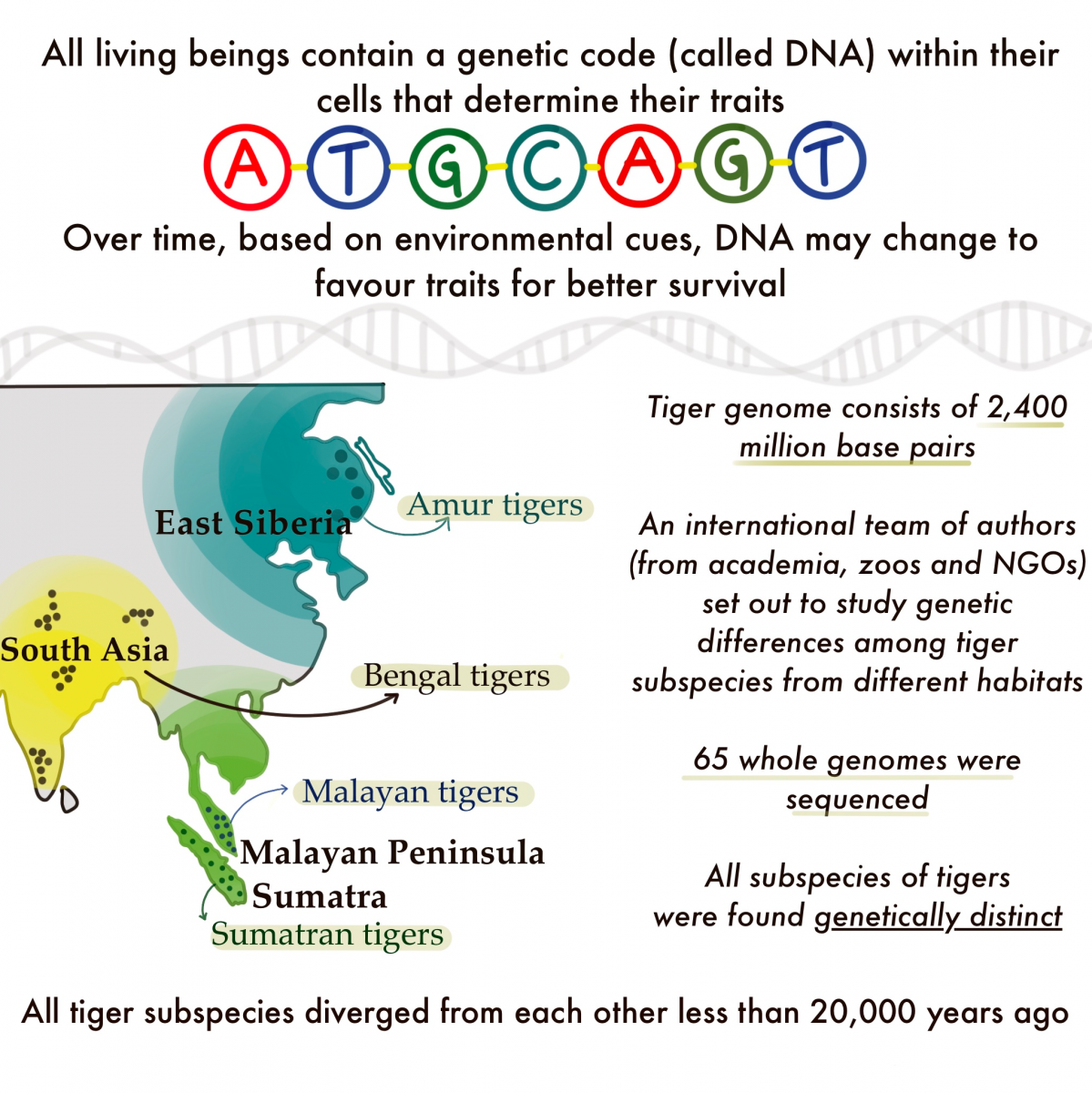
The team sequenced whole genomes from 65 individual tigers from four subspecies, with a specific aim to enhance genomes from wild tigers in different habitats in India. They used these data to conduct a variety of population genomic analyses that quantify genetic variability, and investigate the partitioning of genetic variation, possible impacts of inbreeding and founder events, demographic history (including population divergence) and possible signatures of local adaptation. They found that total genomic variation in Indian tigers was higher than in other subspecies. However, several individual tigers in India had low variation, suggesting possible inbreeding and founding bottlenecks. Tigers from northeast India were the most divergent/different from other populations in India. “Given our results, it is important to understand why some Bengal tigers appear inbred and what the consequences of this are,” says Anubhab Khan, co-first author, NCBS.
The history of tiger populations from across their current range shows recent divergences between tiger subspecies, within the last 20,000 years, which is concordant with a transition from glacial to interglacial climate change and increasing human impacts across Asia. These findings are in sharp contrast to an earlier study by ShuJin Luo and others in 2018 that suggested much older divergence times (https://www.cell.com/current-biology/pdf/S0960-9822(18)31214-4.pdf). The recent divergence between populations will need to be investigated further with expanded datasets and analyses of more tiger genomes. The data and analyses also suggest strong bottlenecks in all tiger populations, highlighting the importance of population size decline on the erosion of genetic variation. “Most studies focusing on species of conservation concern use limited numbers of specimens to try to gain understanding into how genomic variation is partitioned. It is clear from our work here and a growing number of other studies that it is crucial to increase our sampling efforts and use caution when interpreting results from limited sample sizes,” comments Ellie Armstrong, co-first and co-corresponding author, Stanford University.
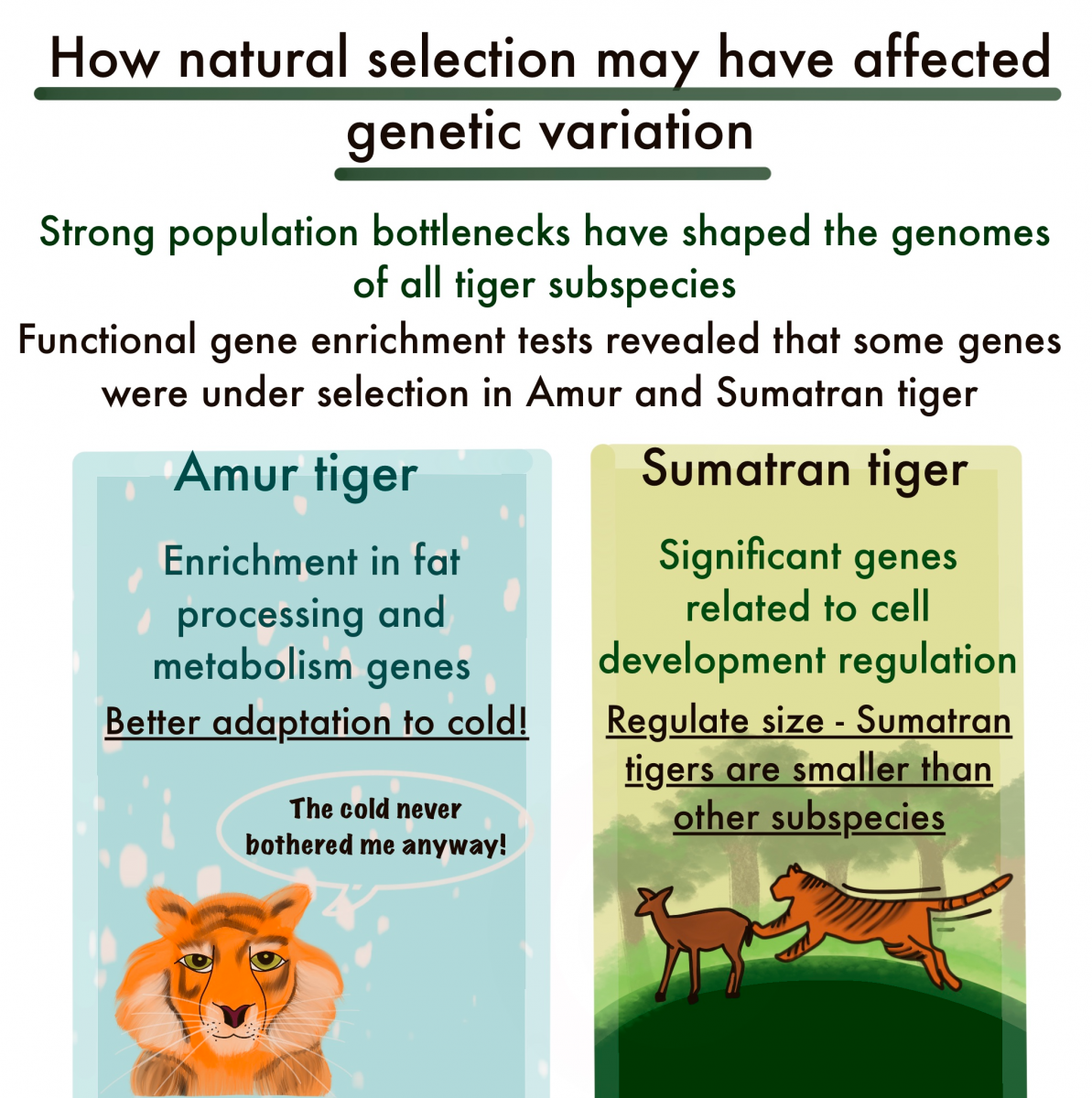
Genomes of tigers in the Russian far east suggest adaptation to the cold, while those of Sumatran tigers suggest selection based on body size. Co-Senior Author Elizabeth Hadly of Stanford University says, “The tiger is an excellent example of the myriad historic events that sculpt species genomic diversity and points to the importance of understanding this diversity as we attempt to stave off extinction of our most precious species on Earth. While some populations demonstrate the importance of adaptation to local conditions, other evidence suggests that particular populations may suffer the effects of climatic change in the Anthropocene.” Such information will be critical to the success of genetic rescue efforts, which should take local adaptation into consideration.

‘I have worked on Indian tiger genetic variation for over a decade and always wondered how they compared to other wild tigers. Our study reveals that while the total variation in Indian tiger genomes is high, they have also been dramatically shaped by population bottlenecks. Genomic variation of Indian tigers continues to be shaped by ongoing loss of connectivity. Population management and conservation action must incorporate information on genetic variation. I hope doing so will help India maintain the gains in tiger conservation achieved so far,” says Uma Ramakrishnan, NCBS, co-senior and co-corresponding author.
Illustrations by Pallavi Raj Sharma.










0 Comments