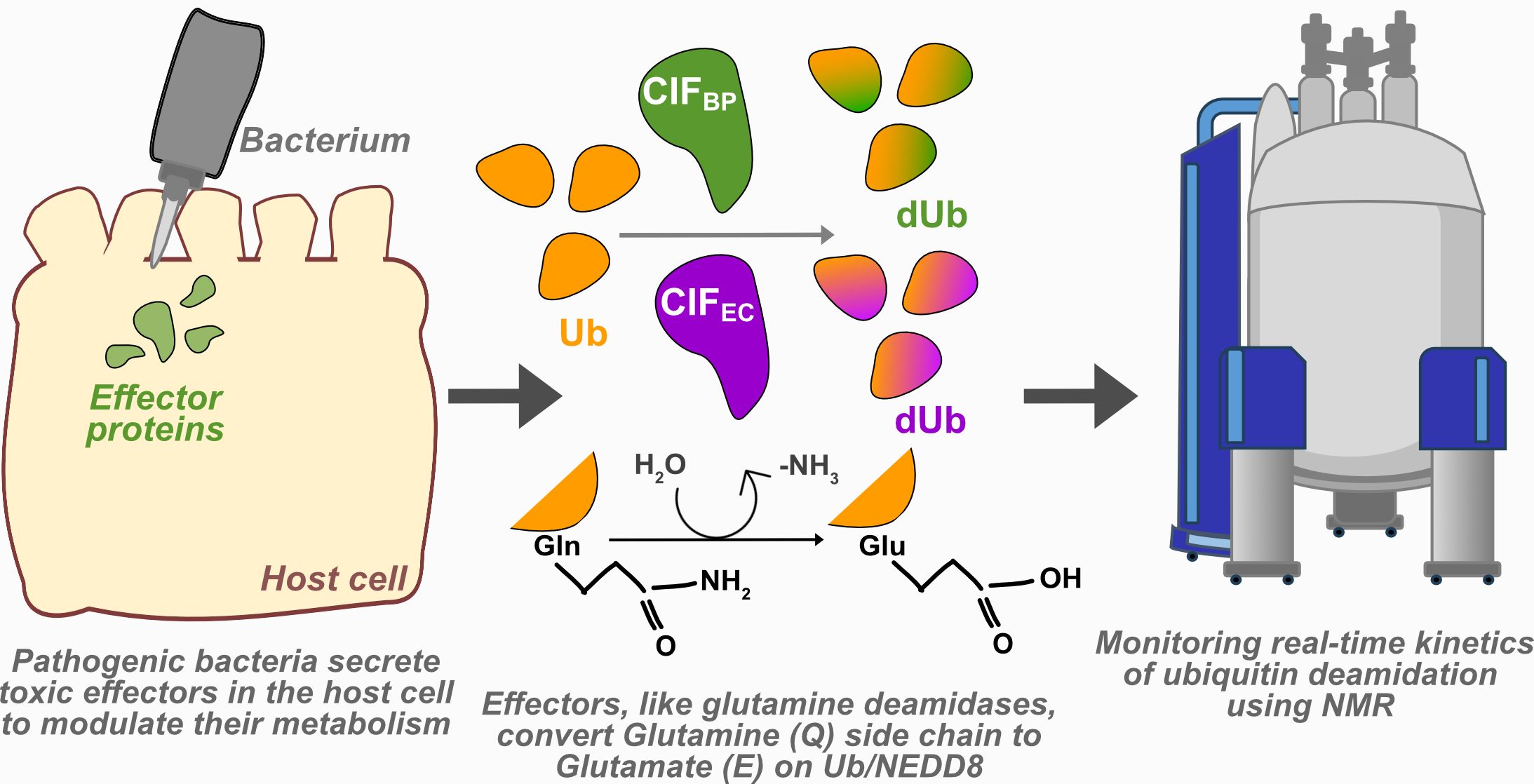
Bacteria have evolved many clever ways to take over human cells during infection. One such trick involves enzymes that chemically modify our proteins to disrupt vital cell functions. A key example is ‘deamidation’ , a chemical change that alters a building block in a protein, subtly but powerfully affecting how that protein behaves. Some bacteria use enzymes called cycle inhibitory factors (CIFs) to do this, targeting important molecules like ‘ubiquitin’ - a protein that acts like a “cellular tag” to regulate everything from immune response to cell division. By modifying ubiquitin, these bacteria can paralyse host defences and interfere with normal cell growth.
Researchers at NCBS, have now developed a new way to watch this sabotage in action. The research team used Nuclear Magnetic Resonance (NMR) spectroscopy, a powerful technique that uses magnetic fields to visualise the changes in molecules to monitor deamidation as it happens in real time.
“NMR is often used to study the shape of proteins, but it is traditionally slow and requires multiple samples taken at different time points, making it difficult to track fast or irreversible reactions like deamidation,” says Dr Rashmi Agrata, the lead author of the study. “The team overcame this limitation by using a faster version of multi-dimensional NMR called BEST-HSQC, which can record data every few minutes without interrupting the reaction,” she added. This innovation allows scientists to continuously follow the chemical transformation of proteins from start to finish, at atomic resolution using a single sample.
Using this method, Rashmi and team compared two bacterial enzymes:
CIFEC from Escherichia coli, and
CIFBP from Burkholderia pseudomallei, the bacterium that causes melioidosis, a fatal tropical disease.
Both enzymes modify the same target - ubiquitin. But the team found that CIFBP acts much more efficiently, working 3 times faster and at 50 times lower concentration than CIFEC. While CIFEC binds to ubiquitin weakly and acts slowly, suggesting a regulatory role, CIFBP appears designed for rapid and aggressive disruption of host systems during infection.
NMR data also revealed that this modification doesn’t distort ubiquitin’s overall structure but changes its local chemical environment - small tweaks that could alter how it interacts with other proteins, with serious consequences for cell regulation.
Beyond explaining how bacteria fine-tune their attacks, this new NMR approach erases one of the biggest limitations of traditional methods: it allows researchers to measure enzyme activity precisely and continuously without destroying the sample. The method can now be applied to study other protein modifications or screen for drugs that block bacterial enzymes.
“This study doesn’t just show how bacteria sabotage our cellular machinery. It lets scientists watch it happen in real time at high-resolution. By making NMR faster and more adaptable, the researchers have opened new doors to studying molecular events that were once too fleeting to catch,” says Dr Ranabir Das, the principal investigator of the study.
Link to the study: https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/mrc.70045?af=R









0 Comments