Scientists from the National Centre for Biological Sciences (NCBS), Bangalore, have developed a test to detect the presence of Merkel cell polyomavirus (MCV) in Merkel cell carcinomas.
A majority (~60-80%) of Merkel cell carcinomas—which are rare and aggressive skin cancers—are caused by MCV, while the rest are caused by long-term ultraviolet light exposure. Usually, patients who suffer from this virus-caused cancer have a better chance of survival, and may benefit more from MCV-targeted therapies than those who contract the cancer because of ultraviolet light exposure. Therefore, detecting the presence of these viruses in Merkel cell carcinomas will be of great diagnostic value not only for prognosis, but also for therapy.
Unfortunately, currently available techniques for detecting MCV in these cancers are expensive, not very sensitive or accurate, and require specialized training and equipment.
The new technique to detect MCV in Merkel cell carcinomas, developed by Dr Reety Arora, Prof Sudhir Krishna, and team at NCBS, can potentially be crafted into a diagnostic tool that will be easy-to-use, less expensive than current methods, and highly sensitive. The results of their work—which is a preliminary, proof-of-principle evaluation for the diagnostic method—have been published in the journal Frontiers in Molecular Biosciences.
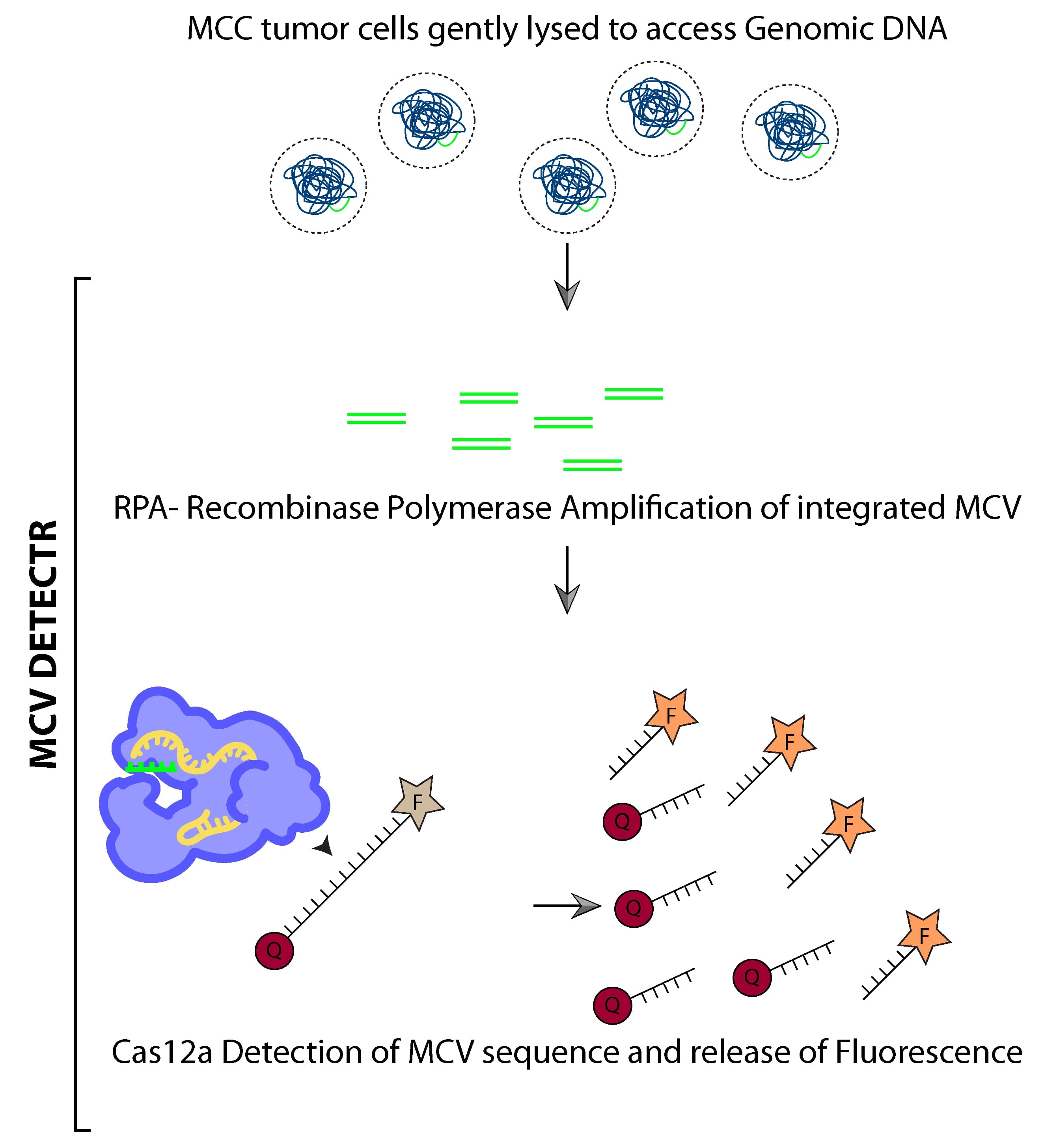
The diagnostic method uses a technique based on a fairly recent application of the genome editing technology, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas system, that was developed from bacterial immune systems. The aptly named DETECTR (DNA Endonuclease Targeted CRISPR Trans Reporter) system has already been successfully applied as a diagnostic to detect the human papillomavirus in patient samples. Based on this system, the researchers have developed an MCV DETECTR.
The MCV DETECTR uses just 3 components—an identifier, a switch, and a reporter—to detect MCV presence in DNA samples of Merkel cell carcinoma tissue. The ‘identifier’ is a guide RNA, a molecule specially designed to recognize and bind to MCV-specific DNA sequences. The ‘switch’ is Cas12a, a DNA-cutting enzyme attached to the guide RNA, and the ‘reporter’ component consists of single stranded DNA (ssDNA) tagged with fluorescent molecules.
If a sample contains MCV DNA, the guide RNAs in the MCV DETECTR system ‘identify’ the target DNA by binding to it, and ‘switch on’ the attached Cas12a. The Cas12a enzymes not only cut the viral DNA at or near the target sequences, they also develop the ability to indiscriminately shred any ssDNA molecules they come across. Now, the ‘reporter’ component of the MCV DETECTR assay comes into play. When attached to the ssDNA, the fluorescent molecules remain ‘quenched’ and do not fluoresce. But as Cas12a begin to destroy the ssDNA, the fluorescent tags are released and start to fluoresce. By measuring this increase in fluorescence, one can detect the presence of, and even quantify the MCVs present in the sample.
To further increase the sensitivity of this system, an additional ‘amplification’ step was also added. In this step, many copies of specific viral DNA targets were generated through a technique known as RPA (recombinase polymerase reaction), which is a simpler, lower-cost, and more rapid alternative to the traditional PCR (polymerase chain reaction), the usual method used to create such copies. With the addition of this amplification step, the MCV DETECTR system can detect MCV in femto-molar quantities.
“Our future plans include developing this as a diagnostic and hopefully in a colorimetric format,” says Dr Reety Arora, a Wellcome Trust-DBT (Department of Biotechnology) postdoctoral fellow, who is the first and corresponding author of the publication on this diagnostic method. Since a colorimetric test would use an indicator that is visible to the naked eye, it will eliminate the need for a fluorescence reader to see the test results. “This way, we won’t need any special equipment or training to perform the test and the test will be easy to use in clinics,” she adds.









0 Comments