Wheat, rye, and barley are some of the oldest grains cultivated by humankind. Staple in many parts of the world, these grains are also infamous for containing gluten. People suffering from celiac disease (CD), have severe immune reactions to gluten, which affects their quality of life.
Those with CD have specific variants of the human leukocyte antigen (HLA) gene, HLA DQ2.5 or DQ8. The HLA protein is known to play a crucial role in the adaptive immune response in humans. Gluten is broken down on consumption into smaller chains of amino acids or peptides. The binding of these gluten peptides to either HLA DQ2.5 or DQ8 can lead to the immune system attacking healthy intestinal cells. This eventually damages the intestinal lining and could lead to an inability to absorb nutrients.
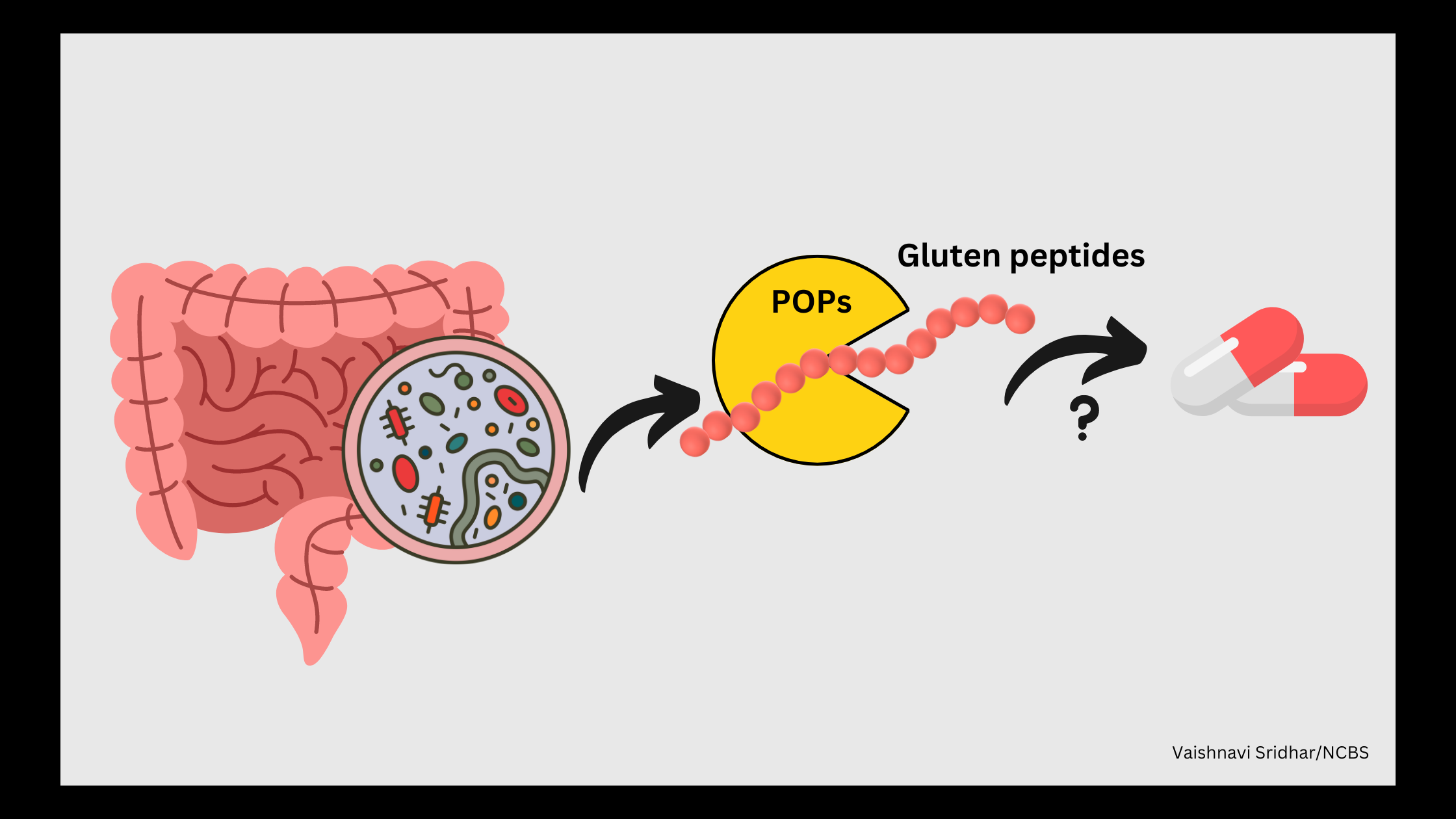
A possible therapy for CD could be to prevent gluten peptides from binding to HLA DQ2.5 or DQ8. Gluten peptides are rich in proline and cannot be broken down by gastric enzymes easily. However, enzymes that degrade proline-rich peptides could be used as potential therapeutics.
Prolyl oligopeptidases (POPs), which are widely distributed in all kingdoms of life, have been found to break down proline-rich peptides. Previous studies have shown that microbial POPs could potentially break down gluten and could be a safe source of therapeutics. However, POPs in microbes and, particularly those from the gut microbiota, are not very well characterised.
A recent study from the National Centre for Biological Sciences, led by senior professor, Prof. R. Sowdhamini set out to characterise the POPs found in the gut microbiota to gain an understanding of their therapeutic potential in alleviating CD related symptoms.
“Current treatments for CD include avoiding gluten-containing foods, consuming probiotics and engineered enzyme-based therapeutics. However, the role of naturally found gut microbiome POPs in degrading gluten peptides has not been explored before”, says Prof. R.Sowdhamini, the corresponding author of this study.
The authors used computational methods to search for gut bacterial genes that fit with the mathematical models of the POP sequence. “Our search identified 6 POP sequences from the bacterial groups; Bacteriodetes, Proteobacteria, and Firmicutes, which are found predominantly in the gut microbiome. We used these POP sequences in subsequent experiments”, says Soumya Nayak, a first co-author of this study.
Using the sequences of these POPs, the researchers predicted their 3D protein structures. They analysed the structures and found structural similarities with a previously well-characterised bacterial POP. They found that the POPs from the gut microbiota contain catalytic domains that are implicated in enzymatic activity.
The authors probed these domains and identified a conserved active site, required for enzymatic activity, and stabilizing residues. These findings hint that these catalytic domains of the POPs could have a potential role in degrading gluten peptides.
POPs have the potential to be highly effective as therapeutics if they can securely bind to gluten peptides without detaching. Moreover, for therapeutics to be successful, they must remain active in the acidic conditions of the stomach and the neutral environment of the intestine. “The ideal therapeutic would break down the gluten peptides in the stomach or at best would sequester these peptides and prevent their interaction with the CD specific HLA in the intestine”, says Dheemanth Reddy Regati, a first co-author of this study.
Using computational methods, the authors found stable interactions between the POPs from the gut microbiota, and gluten peptides. To test the stability of the protein-peptide interaction, the research team simulated the stomach environment. For this, they used the POP sequence from the Prevotella genus, the most prevalent genus in the Indian gut microbiome. They found that the Prevotella POP and gluten peptides interact stably in the simulated stomach environment.
This study highlights a potential therapeutic role for gut microbiome POPs in alleviating CD symptoms. The study further sets the stage for more experiments and functional assays that would help determine the efficacy and applications of the gut microbiome POPs in degrading gluten peptides.
This study was made possible by funding from NCBS-TIFR, DAE, DBT, DST-SERB and IBAB.









0 Comments