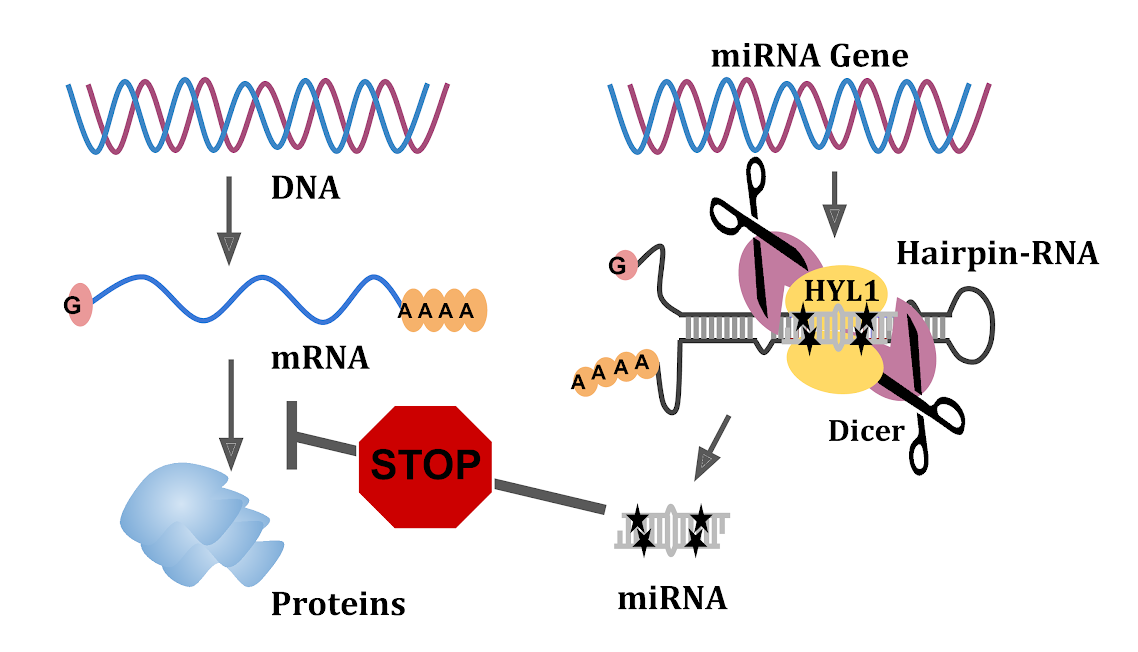
Image: miRNAs stop specific mRNAs from making proteins. This study shows how specific sequences (shown in black stars) in the hairpin loops from which miRNAs originate are essential for the selection of miRNA regions.
Proteins are the major workhorses of any living organism and their availability affects all cellular processes. Organisms should be able to control the levels of proteins quickly in order to respond to any stimulus. This control is achieved by regulating the flow of information at different levels. microRNA (miRNA)s are small regulatory RNA switches of 20-22 bits that are one of the key players. miRNAs control the flow of information from RNAs to proteins by controlling messenger (m)RNA levels. By doing this, miRNAs control which proteins should be made when and where, and at what levels, in a given cell. This is routinely achieved by drastic measures such as by cutting down unwanted and extra mRNAs instead of letting them help in making proteins. miRNA-mediated regulation is important for plants to control all aspects of growth and development, such as during initiation of flowering, development of leaves, control and distribution of hormones to mediate stress responses.
How are these regulators made?
miRNAs are made from long RNA precursor molecules usually with hundreds of digits to fold back to form hairpin-like (stem-loop) structures so that a cutting enzyme can generate small bits of these RNAs. A nicely named protein called Dicer (the one which cuts RNAs) precisely cuts long RNAs, but it is sort of a blind protein without looking for anything other than hairpin-like structures. It was not known how exactly Dicer the cutter knows precisely to select one RNA region from the hairpin-like RNA to make one bit of miRNA.
What has the NCBS team found?
NCBS team has found cryptic sequence signatures in the miRNA region on the precursor. miRNA region has higher levels of two of the four nucleotides (G and C) while the rest of the precursor has higher levels of other two nucleotides (A and U). This higher GC composition was maintained in a majority of plant miRNAs by having a position-specific bias for these nucleotides. This bias for specific digits was seen in miRNAs that are present across all classes of plants from mosses to flowering plants. NCBS team used a variety of techniques to show which digits among the 21 must be G or U for generating miRNAs. They were even able to turn a poorly made miRNA into highly abundant miRNA by incorporating these sequence signatures. Since Dicer is blind, the NCBS team looked for proteins that can help is recognizing such signatures and identified an RNA binding protein partner of Dicer which helps in the selection of sequence signatures.
Is there any use for this finding?
In order to study the function of a given gene, one way to experiment is by removing or reducing the protein made by this gene. In plants, an attractive method called artificial miRNA technology is routinely used, and in this technology, one can design an artificial miRNA. Since this technology did not take into account what the NCBS team identified, one has to use this technology on a trial and error basis. NCBS team has also shown that by incorporating specific digits, this technology can be used directly with high efficiency.
“Our research reports a mechanism which explains the generation of all abundant miRNAs in plant cells. Previous efforts from few laboratories including ours found mechanisms applicable only to few miRNAs” - says Anushree, main author of the paper published in Nucleic Acids Research. The published paper can be accessed here.







0 Comments