Swapping long-drawn data searches for ingenuity: 3DSwap
Scientists studying Alzheimer's, amyloidosis or similar diseases caused by malformed proteins won't have to sift through pages of literature anymore. Searching for information including the structures of proteins involved in diseases like these is now easy, thanks to 3DSwap, an online database developed by a team of researchers at NCBS. 3DSwap is a freely-accessible database of proteins recorded in diseases caused due to protein malformation.
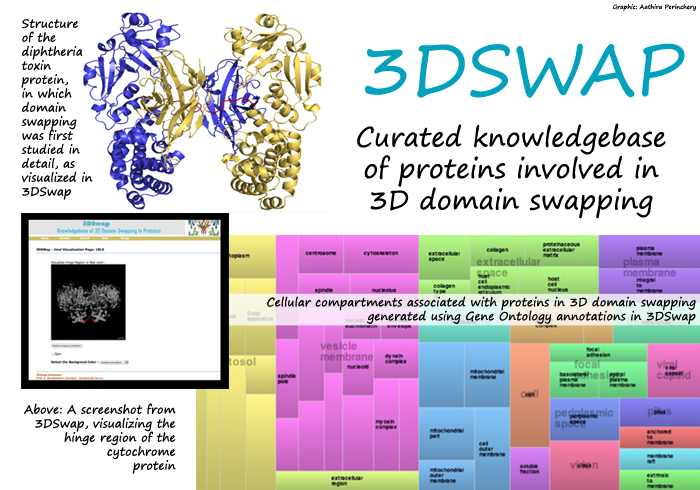
A protein is made up of a chain of amino acids. The function a particular protein performs depends on how these amino acids are spatially arranged. In a complex protein, amino acids can be further clustered into two or more semi-independent parts called domains. Domains are important in protein evolution, since they can be exchanged or swapped within a single protein or between many proteins to form new proteins: a phenomenon called domain swapping. Sometimes identical proteins swap similar domains to evolve more complex proteins in a process called 3D domain swapping. Studies showthat 3D domain swapping plays a vital role in regulating both the physiological and biological functions of these new proteins.
But sometimes 3D domain swapping can go horribly wrong. Malfunctioning proteins formed as a result cause diseases. Studies reveal that abnormal changes in protein structure during 3D domain swapping result in medical conditions like prion and Alzheimer's diseases, where insoluble proteins aggregate abnormally. Identifying the proteins and their changed structures as a result of 3D domain swapping, therefore, plays an important part in studying the causes of some of these diseases.
That's what a team of scientists at NCBS have contributed their bit to. In a study published in August 2011 in the journal Database, five scientists - Khader Shameer, Prashant Shingate, S. C. P. Manjunath, M. Karthika and Ganesan Pugalenthi - with principal investigator Ramanathan Sowdhamini - developed the protein database 3DSwap. The database provides information on 293 proteins involved in 3D domain swapping. The authors compiled data through several stages of analysis. They first used keyword searches in Protein Data Bank (an online repository for three dimensional structures of proteins) and PudMed (an online collection of scientific articles related to biomedical literature). To accomplish this, they wrote their own codes and programs. They traced each protein in the resulting list to their respective scientific articles. From these articles, the authors curated information about the proteins and their structures. In some cases, they extracted information hidden in figure legends and tables: a lot of painstaking work. Says Shingate, one of the authors, "I personally went through nearly 800 abstracts and 150 papers in detail." The team combed through 1200 abstracts and 400 full-length scientific papers in total.
Thanks to all the meticulous work that went into its creation, 3DSwap is a comprehensive collection of information about proteins involved in 3D domain swapping. The information listed for each protein includes its sequence structure, swapped and 'hinge' regions, protein structure with its chemical bonds and the specific functions each protein performs. Users can visualize the structure of each protein using an interactive interface. "There was no single quantitative method to define what percentage of the protein structure may have been affected by 3D domain swapping," says Shameer, the first author of the paper. To help researchers understand this, the authors defined new concepts like 'extent of swapping', which the database lists for each protein. Users can also browse for protein structures using keywords and download data as a textfile from 3DSwap.
"3DSwap is an amalgamation of intense literature survey using text mining, graphical structural analysis of proteins and computer algorithms. So the curation of this database was quite challenging," says principal investigator Sowdhamini. Her team is now updating the list of proteins in 3DSwap.
"We believe this platform will be ideal for large-scale studies targeted to understand swapping and various biological and clinical aspects associated with this unique structural phenomenon," says Shameer. According to the authors, 3DSwap will be a useful tool for "experimental biochemists, structural bioinformaticians, biologists and clinical investigators who intend to study the diverse mechanisms, functions and diseases mediated by 3D domain swapping."
The paper can be accessed at http://database.oxfordjournals.org/content/2011/bar042.abstract.
The database 3Dswap can be accessed at http://caps.ncbs.res.in/3dswap/.
Comments
Please find the 3DSWAP
Post new comment