Beyond Microscopy: Nanoscopy!
But at the 2nd Bangalore Microscopy Course that ran February 21-28 at NCBS, participants drawn from India and other parts of Asia learned how to break the 200nm barrier with a range of new techniques described under the umbrella term of “super resolution microscopy”. These techniques, referred to by a range of acronyms – e.g. SIM, STED, STORM and PALM - increase the resolving power by at least a factor of two and up to 50 times or more, in some cases even allowing molecules of several nanometres to be resolved. The revolutionary potential of “fluorescence nanoscopy” was recognised by Nature Methods in 2008 by awarding it the Method of the Year. The Microscopy Course attendees not only learned about the techniques first hand from the heads of the labs where several of these techniques were first applied – but also got hands-on experience in using the technology itself.
“Super resolution microscopy removes obstacles faced in light microscopy, which has been employed for more than a century. It provides biologists a chance to revisit various processes within the cells at higher resolution and arrive at new inferences that can revolutionize biology,” says Bo Huang assistant professor at University of California San Francisco, and major collaborator with Xiaowei Zhuang, developer of the STORM technique. It is now being used to study the organization of chromatin, proteins in cellular membranes, and synapses of neurons, to mention a few significant areas of active study. The upheaval in super resolution microscopy, as applied to biology, started with the development of STED (stimulated emission depletion) by Stefan Hell at Max Planck Institute of Biophysical Chemistry, Gottingen. Eric Betzig and Harald Hess at Janelia Farm, Howard Hughes Medical Institute developed PALM (photo activated localization microscopy) to achieve temporal and spatial resolution in molecules that are 25 nanometers apart. In order to understand the behaviour of single molecules, STORM (stochastic optical reconstruction microscopy) was developed by Xiaowei Zhuang’s laboratory at Harvard University. Satyajit Mayor, dean of NCBS already utilizes STED, to solve puzzles in cell biology, in particular the structure of “lipid rafts” of clustered proteins in the plasma membrane.The tightly-packed course laid special emphasis on super resolution microscopy and the participants felt privileged to learn about these cutting edge techniques in microscopy. “The didactic lectures were superb. The structure of the course was also well planned,” said Arpan Kumar Rai from TIFR Mumbai. The friendly atmosphere typical of NCBS events was also much appreciated: “The extremely informal set-up between instructors and students made it a dynamic workshop”, said Dhananjay Huilgol, TIFR, Mumbai.
A demonstration, using the same cell, of the increased resolving power of super-resolution microscopy
Traditional fluorescence microscopy reveals the general location of the fluorescently labeled membrane protein farnesyl within a cell. This photo, and explanatory text (and the two photos/text below): modified from photos provided by kind permission of Dr Harald Hess, Janelia Farm, Howard Hughes Medical Institute, Ashburn, Virginia.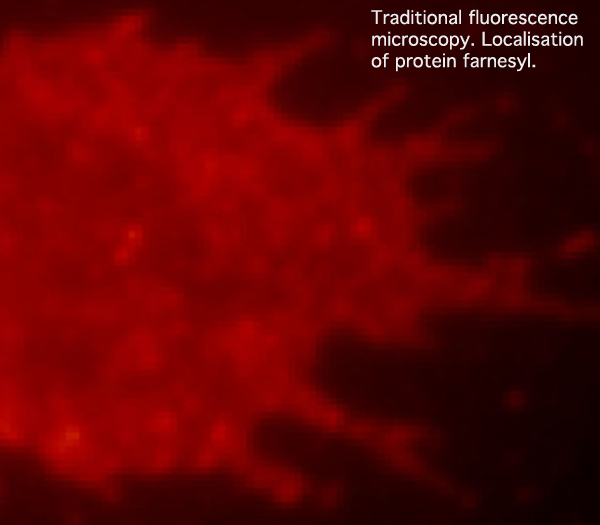
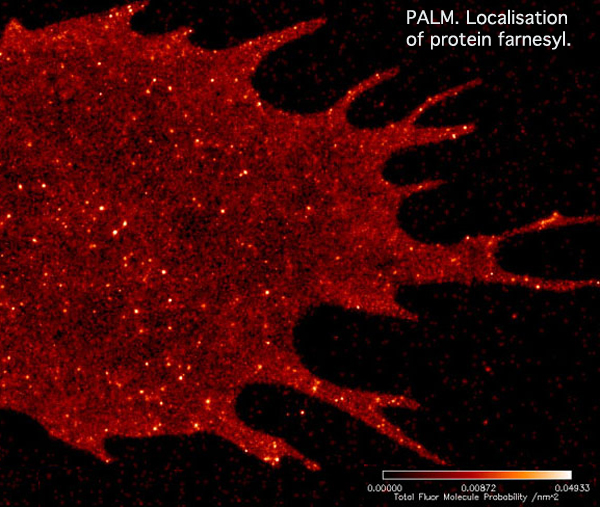
With PALM imaging, the two dimensional distribution of the labeled membrane proteins becomes much clearer. However, it is impossible to determine the vertical position of the fluorescent molecules in the flat image.
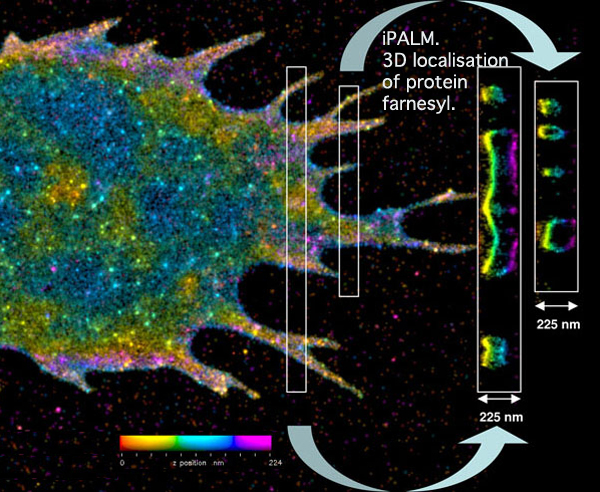
iPALM pinpoints the three-dimensional distribution of the fluorescently tagged membrane proteins. In this image, the vertical position has been color coded, with red molecules being the deepest and purple the highest. Cross-sections of small regions of the image are shown in the white boxes on the right, and reveal two layers of the labeled membrane proteins -- at the top and bottom of the cell.


Comments
Bangalore Microscopy Course
Post new comment