The question of how life arose on earth has always intrigued us. In particular, the question of how unicellular organisms transition into multicellular forms of life, has inspired the work of several scientists across the globe.
Multicellular systems have hallmark features like division of labour and spatial organisation of cells. But how do genetically identical single cells transform into groups of cells exhibiting these hallmark features? While studies have covered the genetic and evolutionary aspects involved in such transitions, very few have determined organising principles that determine the development of cell communities.
Researchers from the Institute for Stem Cell Science & Regenerative Medicine (inStem), Bangalore and the Simons Centre for the Study of Living Machines, at the National Centre for Biological Sciences (TIFR), explored this question. Working with yeast cells as a model system, they have shown for the first time, how cells exhibiting very different properties arise and then organise themselves into groups or patterns, based on the types of metabolism that can be carried out in the given conditions.
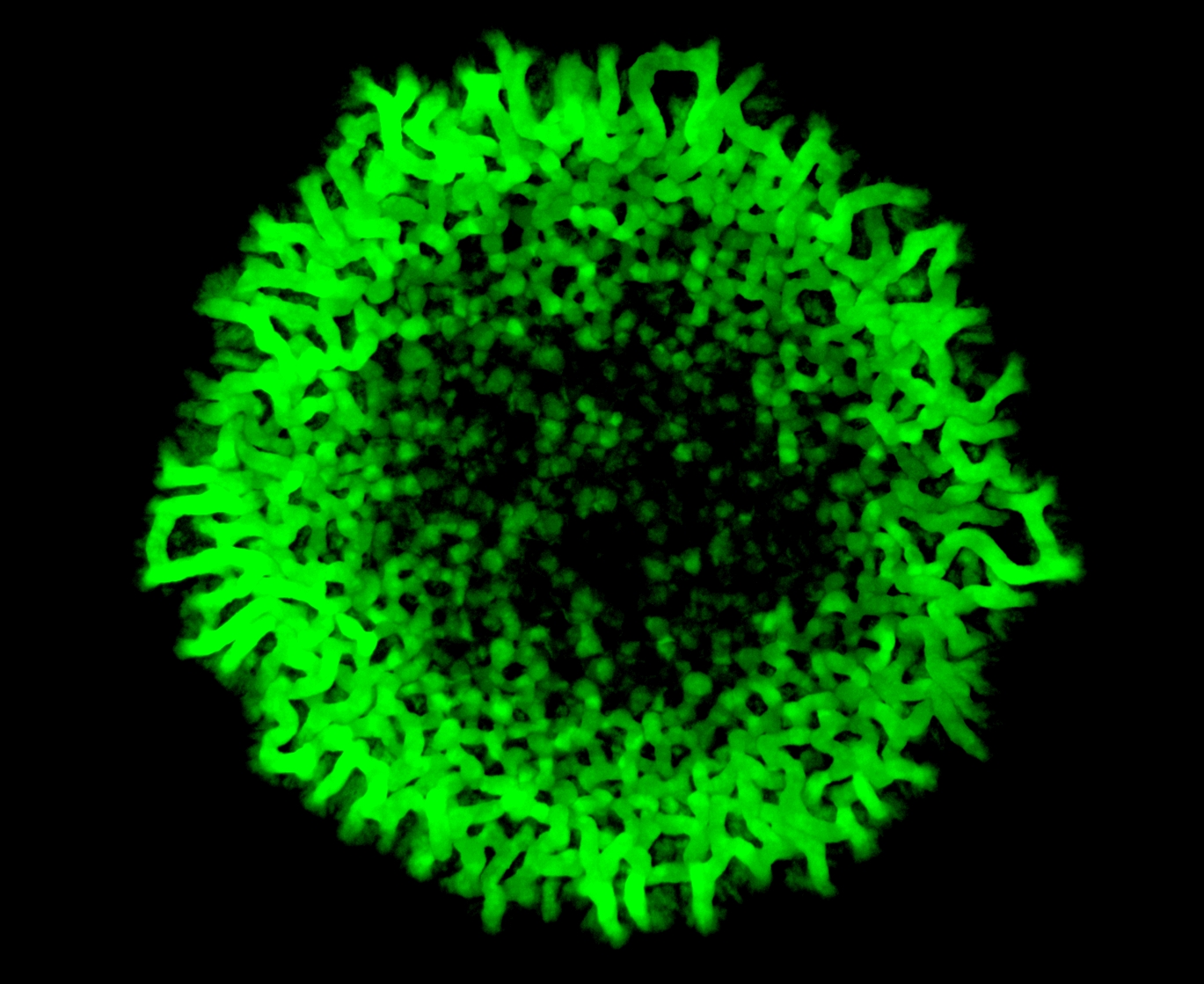
Dr. Sriram Varahan, the lead author of this paper, explains that when cells have very little glucose, they try to make glucose from other sources of carbon. When they do so, one molecule they make is trehalose – a simple ‘disaccharide’ that resembles sucrose (table sugar). At first, all cells in the community carry out only this biochemical process. Interestingly, over several days, the researchers observed that cells which looked different emerged around the periphery of these yeast colonies, and were organised with very clear patterns. These new cells grew much faster than the original state cells. The researchers realised that the new cells were in an entirely different metabolic state. Instead of producing trehalose, the cells in the new state exclusively used up trehalose through another metabolic pathway, and turned this sugar into food. Effectively, these cells were entirely dependent on the original population of cells making trehalose.
Vaibhhav Sinha, a graduate student with Dr. Sandeep Krishna at the Simons Centre at NCBS, decided to model this transition, using coarse-grain computer simulations in a hybrid model. Vaibhhav explains “we were interested in looking at how local changes, decisions, and interactions can influence or give rise to recognisable features at the colony level”. The model corroborated Sriram’s experimental observations, suggesting that for the cells in the new state to emerge, the original group of cells produce a resource; which was used up by the newly emergent cells in the different state. The model can simulate the pattering and organisation of cells in different state.
This study is a conceptual advance, explaining how rules from biochemistry and physics can define how cells showing different properties can emerge, and then organise into patterned, interdependent communities. Sriram speculates that the co-existence of cells in different metabolic states can help the yeast colony, as cells with different properties lend new, adaptive advantages to the entire community. Sriram also speculates that this study helps us understand how ‘multicellularity’ can emerge in yeast-like systems. Cells start as simple aggregations, but then develop both division of labour and mutual interdependence and distinguish themselves from each other. Dr. Sunil Laxman, the corresponding author in this study elaborates, “This concept is of wide interest to people studying infectious microbes and biofilms, as also cancer, where tumours have organised groups of cells, with different properties, making them hard to kill.”
The publication titled, ‘Metabolic constraints drive self-organization of specialized cell groups’ was recently published in the journal, eLIFE.
Dr. Sunil Laxman can be reached for further correspondence at sunil@instem.res.in.
This work was supported by a Wellcome Trust-DBT India Alliance, Department of Biotechnology (Govt. of India), and institutional support from inStem, NCBS-TIFR, and the Simons Foundation.
Cover image: Spatially restricted gluconeogenesis activity across a yeast colony.
Image Credit: Sriram Varahan









0 Comments