Along with the those on the frontlines, thousands of researchers around the world are working to identify the right approach to tackling the COVID-19 pandemic from various angles and disciplines. The search for effective treatment options and vaccinations for the SARS-CoV-2 has occupied scientists and medical researchers in recent months in an unprecedented manner. NCBS researchers were quick to join in this hunt, and Prof. R. Sowdhamini’s group began with a plan to identify compounds that can be repurposed in the battle against the SARS-CoV-2.
In early March, the team began their search with 3,30,000 small molecules off the FDA-approved drug database, and others of natural origin, mostly from plants. They were trying to narrow down the elusive needle in the haystack of solutions. With their computational approach, they have achieved a huge reduction in molecules, and only one in 10,000 molecules have passed their exhaustive computational pipeline, which saves time, money and effort in experimental examinations.
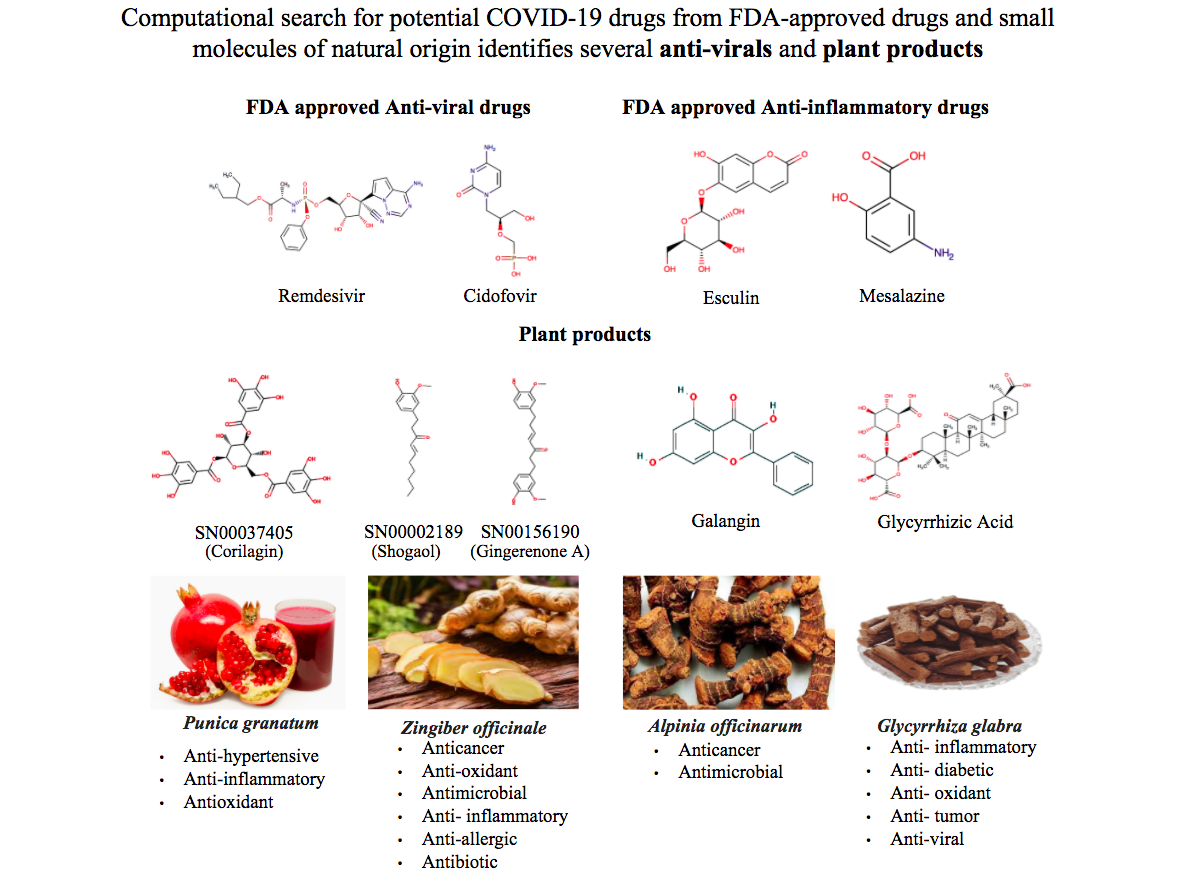
They suggest 33 small molecules which could bind to one of the SARS-CoV-2 proteins, NSP1 and inhibit it. The molecules they have identified are those which can be repurposed safely from FDA-approved drugs or natural sources, which ensures that significant time can be saved on toxicity tests. The shortlisted molecules can be taken forward for wet-lab validation, both for binding and to reduce viral application. The lab’s research will be published in the Journal of Biosciences.
The NSP1 protein became the target of Prof. Sowdhamini’s group, as a protein in the SARS-CoV-2 which works very insidiously against the people who are infected while helping to keep the virus immune to the body’s defences. Prof. Sowdhamini says, “The choice of target protein, NSP1, is both unique among research at this time, and important based on the role the NSP1 protein plays. It stalls host gene expression, could degrade host mRNA, and protects viral (self) mRNA from degradation. It is a great interactor with other proteins as well.” The researchers searched for NSP1 interactions against a large set of molecules, and their stringent measures narrowed down the list to 33 molecules with stable and strong interactions with this protein.
The team worked on a series of processes to perform the virtual screening, including taking into account the ways the molecule would interact to inhibit the protein, based on the protein structure. The virtual screening they used requires computational docking algorithms for protein-ligand interactions, where shape and charge complementarity are considered. “Scores were provided depending on the strength of favourable interactions (such as hydrogen bonds) and the presence of unfavourable interactions (such as short contacts). More negative the scores, higher the chance for that molecule to be a potential inhibitor for the NSP1 protein,” says Abhishek Sharma, the first author of the paper.
After two rounds of filters, the top-ranking molecules were considered. “These docking runs took approximately 20 days of computing time, followed by analysis and literature survey,” adds Sharma. The processing was computationally intense, which was followed by molecular dynamics simulations to yield the small molecules with the most stable interactions with the NSP1 protein. “We find out if the small molecule or ‘ligand’ stays on with the protein and does not fly away during our simulations,” says Vikas Tiwari, who performed these simulations.
In the virtual screening with NSP1, FDA-approved anti-viral compounds like Remdesivir and Edoxudine are promising. Esculin and Acarbose which are not anti-viral drugs, but are used as anti-inflammatory and antidiabetics respectively could be used as potential inhibitors. In addition to these, the 33 short-listed molecules which form stable interactions include lactose, Glycyrrhizic acid from Liquorice (also referred to as Athimathuram or Yastimadhu), Corilagin from pomegranates, Galangan from Blue Ginger (also referred to as Galangal or Sitharathai), and Gingeronone and Shogaol from both Ginger and Blue Ginger.
“We started this work during early March and it is great to see that we could recommend a set of promising small molecules to bind to NSP1. But, what will make us happier is if some of these ligands could be useful to combat COVID-19,” says Prof. Sowdhamini.
With these computational efforts, the team has helped pave the way for more investigations of shortlisted molecules. And with the careful selection of the molecules, and other work of this nature, the strides towards a potential preventative and treatment are growing greater every day.
Image credit: Abhishek Sharma








0 Comments