Translation, the process by which information from messenger RNA (mRNA) is decoded to build proteins, is a central process to all of life. The nuts and bolts of the translation machinery are among the first concepts biology students learn. Yet, what is not apparent to many is that the components of translation can be diverse across species. Gaurav Diwan and Deepa Agashe from the National Centre for Biological Sciences (NCBS) trace the history of some of the key components of bacterial translation, to understand their role in the evolution of bacterial translation.
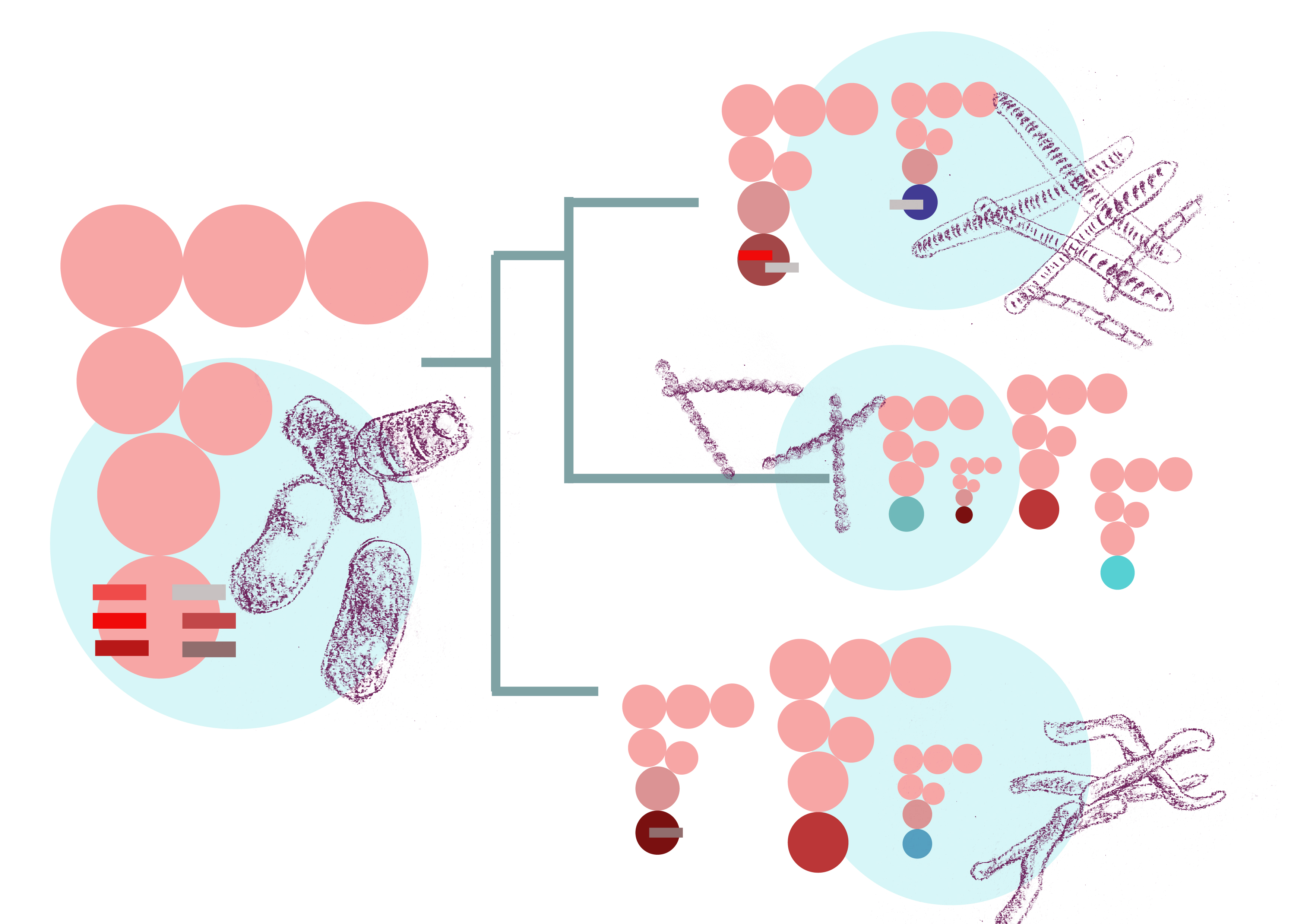
The transfer RNA, or tRNA, is the molecular bridge that matches codons—units of genetic code—on the mRNA with the amino acids they code for. A total of 61 codons encode the 20 different amino acids. One would expect that there are 61 types of tRNAs to match. Bacteria, however, show large diversity in their tRNA repertoire. Some have about 45-46 types of tRNA, while others have just 9!
How do bacteria carrying only a few types of tRNA decode all 61 codons? A set of enzymes called modifying enzymes chemically modify bases in some tRNAs, causing them to “wobble base pair" with codons that they normally cannot bind with. What this means then is that with a much smaller subset of tRNAs, the cell can decode a larger number of codons.
Different bacteria have different modifying enzymes. In their latest paper in Molecular Biology and Evolution, Diwan and Agashe have investigated, for the first time, how this diversity has emerged over time in bacteria and how changes in the wobble enzyme machinery are associated with changes in the tRNA repertoire.
The researchers used a phylogenetic tree of almost 1100 bacteria. At the tips of this tree are presently known bacteria for which complete genome sequences are available. For many of these species the tRNAs and their sequences are also known. The modifying enzymes were the missing piece. Diwan scanned across the 1100 genomes to identify previously known modifying enzymes, catalog the different enzymes found in each bacterium, and spot gains or losses of these enzymes over time. Some very nice patterns were immediately apparent—entire modification pathways are missing in specific groups of bacteria while some pathways are found across all bacteria in the phylogeny.
Armed with these data, they sought to test a simple hypothesis: if a given type of wobble base pairing exists in a bacterial species, it could make do with a reduced set of tRNAs. Conversely, if this wobble base pairing does not exist in another bacterium, that bacterium would have more/different types of tRNA. Diwan combed through the evolutionary history of bacteria to test if the loss of modifying enzymes correlated with a larger tRNA repertoire. By comparing the tRNA repertoires between sister clades—closest relatives in the evolutionary tree—where major gain or loss events had been identified, they found that the hypothesis is largely borne out: when the wobble is lost, there is an increase in the tRNA repertoire.
What causes this loss in wobble? The researchers find that shifts in GC content—the percentage of guanine (G) and cytosine (C) bases in the genome—are strongly correlated with gain or loss of modifying enzymes. It is not known why GC shifts occur, but these shifts alter the codons that are used to encode genes. Now to decode these codons, a different set of tRNAs are required and the wobble may not be needed. Also once there is an increase in tRNA diversity, the wobble can be lost.
So the reason wobble is lost is not because it offers a fitness benefit. “Our analysis gives a glimpse of the translation machinery of the common ancestor of all bacteria, which lived billions of years ago. It probably did not have a full set of tRNAs, resulting in strong selection to maintain its wobble enzymes. Later, as the tRNA diversity in bacteria increased because of changes in GC content, selection maintaining wobble enzymes was weakened in some bacteria. So, we’re suggesting that the genetic code itself was shaped along with wobble,” explains Agashe. Other projects in her lab seek to understand the causes underlying GC shifts, which are turning out to be important drivers of many aspects of bacterial genome evolution.
This work highlights the unexpected diversity and dynamics of the translation machinery of bacteria, and poses new questions about the evolution of the genetic code and genome evolution. “We have just about scratched the surface of the diversity of translation across bacteria,” says Diwan.
The paper titled 'Wobbling Forth and Drifting Back: The Evolutionary History and Impact of Bacterial tRNA Modifications', can be accessed here.
Illustration: Ipsa Jain









0 Comments