Active Actin Asters
A cut finger calls into action a crew of repair cells - macrophages - that crawl amoeba-like towards the wound. To drag itself forward a macrophage repeatedly anchors, then detaches, its advancing membranous fringe, to and from, the surrounding matrix. This feat is just one of many activities made possible by the cell's ability to enrich selected areas of its outer membrane with specialized proteins. For example, the macrophage's moving fringe is enriched in the protein integrin, which can link the membrane to the matrix. How such localized enrichment occurs within the broad expanse of the cell's membrane has remained unclear. Now however a team of researchers at the National Centre for Biological Science and the Raman Research Institute, Bangalore, believe they have the answer. In a recent paper in the journal Cell (Gowrishankar et al., 2012) they have proposed, and tested, a model that, unlike previous models, explains how enrichment could be an active process, and thus controllable by the cell.
The outer membrane is not random
The idea that the cell's outer membrane is not a random mixture of lipids and proteins was popularized by the Finnish scientist, Kai Simons in the 1990s. He proposed that "lipid rafts" could form passively within the membrane, a consequence of the natural tendency of some lipids to prefer, as it were, the company of their own kind. Each lipid raft could also host a bunch of "piggyback" proteins that could connect up to the aggregating lipids. In this otherwise very attractive model, a cell could, however, not specify where rafts would form. Later it was suggested that the cell could solve this problem if it were able to "fence-off" membrane regions within which the rafts were constrained. There was even a perfect candidate for the fences: a loose meshwork of semi-rigid filaments (made of the protein, actin) that lies directly under the membrane (see figure below). Each sector of the actin mesh could help define a holding pen for a microscopic armada of lipid rafts.
The Bangalore group had already provided some of the best experimental evidence that the composition of the membrane is non-random, and that actin plays a part in its regulation. Focusing on one type of piggyback protein (GPI-anchored proteins), their previous work had however raised doubts about fundamental aspects of the lipid raft/actin fences model. They showed that GPI-anchored proteins did not form the kind of clusters conjured by Simon's raft model, but formed nanoclusters and monomers, in a proportion that was inconsistent with any kind of passive aggregation mechanism.
Also, while actin did appear to help these proteins form "nanoclusters" that further congregate into grander "superclusters", the superclusters were profoundly smaller than the sectors of the actin meshwork. Further evidence against a static, fence-like role for actin came from experiments that showed that clustering of GPI-anchored proteins required myosin. This protein is normally involved in dynamic processes in the cell, for example when it teams up with actin and ATP during muscle contraction.
Enter the asters
Aware of the problems besetting the lipid raft model, and the desirability of a cell having control over nanocluster formation, the Bangalore team now adopted some wild and subversive ideas. What if a second type of actin filament - a much shorter, more dynamic form - was interspersed amongst the meshwork actin: How might these behave if they interacted with myosin in an ATP-driven way? What if the formation of each individual nanocluster was not passive but active?
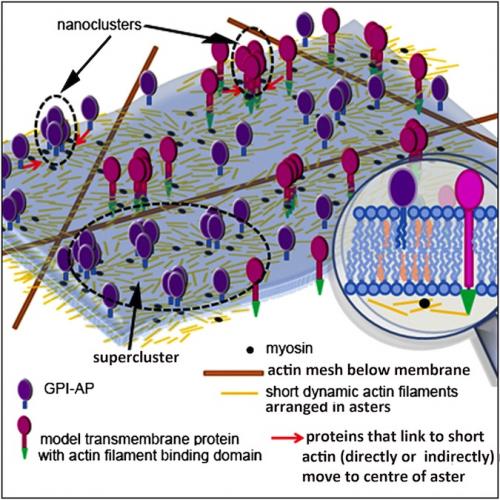
The Bangalore team proposes that actin filaments of a new type - short, and constantly remodelling - are arranged in asters underneath the the cell's outer membrane. If a membrane protein can bind (directly or indirectly) to these filaments, it could be carried towards the centre of an aster, leading to nanocluster formation. A cell could specify where nanoclusters form by promoting aster formation, or by boosting levels of myosin or ATP; the latter help short filaments to organise as inward-directed asters. From Gowrishankar et al., 2012 (edited).
The team now had a nanocluster formation model that, by invoking ATP-requiring interactions between actin and myosin, was active, and not passive. A cell could put its proteins where it wanted.
This was strong, exciting stuff, and it was time to run further with it. First, Gowrishankar and Rao scrutinised all the known behaviours of the GPI-anchored proteins that had been the focus of the team's many previous papers. Their model was able to explain each and every behaviour noted in their earlier work.
In vivo veritas
The experimentalists in the team, Subhasri Ghosh, Suvrajit Saha, Rumamol C. and Satyajit Mayor worked with the theoreticians to "ground-truth" the model: to look for more convincing evidence of what was assumed, and predicted, by the model. Actin filaments of any sort are notoriously difficult to visualise, but even more so when they are as short and as dynamic as the theoreticians had proposed. But the team relishes extreme microscopy. While they could not directly observe the short filaments in their entirety, they were able to reconstruct their movements from traces of their motion as seen down the microscope. These movements were sensitive to drugs that break down, or stabilise, actin and this was just as the theoreticians had predicted.
They also verified two predictions of the theoretical model. They introduced artificial actin-binding proteins into cells, and found that these aggregated as superclusters within the membrane, suggesting they may have been captured by the proposed nanoclustering mechanism. In addition, GPI-anchored proteins within the cell's membrane appeared to exhibit the non-random movements predicted by the model. When these proteins were tagged with a fluorescent dye, the intensity of their fluorescence fluctuated more wildly than that expected for particles moving randomly.
The work, a rich interweaving of theory and experiment, threatens to turn the lipid raft model on its head. Cells can selectively enrich areas within the outer membrane, by specifying where nanoclusters and superclusters form. It also helps explain why previous workers have never been able to replicate, in living cells, the large lipid domains that do form spontaneously in artificial membranes. In the wider view, with a broader group of participating molecules than the previous model, the work suggests more points where nanocluster-dependent activities might malfunction, and at the same time, where interventions could be made. The paper has already spawned a "preview" article in the Cell Press journal, Developmental Cell, a sure sign that fellow workers in the field are excited.
The paper:
Kripa Gowrishankar, Subhasri Ghosh, Suvrajit Saha, Rumamol C., Satyajit Mayor, Madan Rao
Active Remodeling of Cortical Actin Regulates Spatiotemporal Organization of Cell Surface Molecules
Cell, 2012, 149, pp 1353-1367
http://www.sciencedirect.com/science/article/pii/S009286741200582X


Comments
Post new comment