"Independence is a good thing"
Shachi, tell us how you got into the sort of science you do?
I joined my integrated Masters in chemistry, but that was incidental. I became interested in physical chemistry in my final year because I liked the more quantitative aspects of it. During my Masters thesis project I got interested in doing theory and computation. So, that was the direction I decided to pursue when I joined Caltech for my PhD. By training I'm a theoretical physical chemist though I did do a few biochemistry related courses in my undergraduate years. But all through my PhD I was doing very non-biological research, looking at electron transfer at metal surfaces.
It's quite a jump from that to working on proteins.
Yes, I got interested in enzymes because there is a lot of electron transfer going on there. Also, my PhD advisor and others in the group were interested in both electron transfer and solvation dynamics in proteins.
Towards the end of my PhD, I was trying to work out what to do next. As part of the Caltech PhD program, we had to write three proposals - one closely related to the research topic and two in other fields. One of my unrelated proposals was on parallel computation and the other was on protein folding. At that point I decided that I would work on proteins for my postdoctoral research and I applied mostly to people who worked on biological systems, both computational and theoretical. I ended up doing computational work with Prof. Onuchic at UC San Diego. He had started off in electron transfer as well so he seemed comfortable with letting me loose on protein folding problems.
Did you need to bring yourself up to speed with the biology?
Not really, the project involved in vitro protein folding with simple models of proteins, so it was more about learning biological physics. The biology challenge came later, when I came to NCBS.
Do you enjoy moving into new areas?
Yes. You tend to learn a lot, it's always nice, getting a fresh outlook. Sometimes your old ideas work out in the new field and sometimes they don't. But there is always something new to learn. And for the new field, there's always value when people come in without preconceived ideas.
And how long were you at UC San Diego?
Six years. I was starting on something completely new - I had done few molecular dynamics simulations before. I had to develop a new method to work on proteins that were slow folding, so that took a bit of time.
This led to your interleukin papers?
Yes, the folding of interleukin turned out to be very interesting. But the first couple of years just involved making sure that the simulated protein did actually fold and unfold. The method development ended up being the harder part.
Was that a new methodology?
No, it was a modification of an old approach so that it would work for my system. But it also works well for large proteins. Most people are not willing to study proteins that fold on a minute timescale and that was what worked for me.
That is a coarse-grained approach to folding?
Yes.
To some people, simulated protein folding might seem a bit unreal. What would you say has been the single greatest insight that has emerged from computational and theoretical approaches to protein folding?
Outsiders to the field might be misled by the apparent level of contentiousness in the protein folding world. The debates that occur are really just quibbles and we know quite a lot about protein folding.
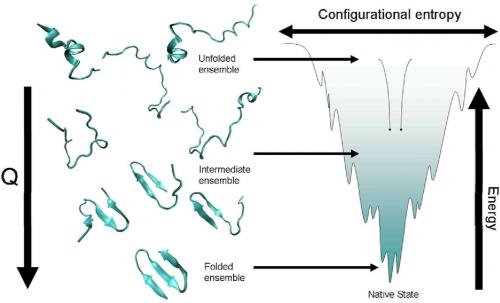
The biggest breakthrough was the energy landscape theory. The big question was how do proteins solve the folding search problem. An unfolded protein chain has a lot of states and if the protein had to search through all these states it would take too long to fold. The theory was then proposed that proteins are not random but special amino acid sequences that had evolved to fold efficiently. This idea is a rather profound way of thinking about proteins. What it led to is an understanding of the energy landscape that proteins traverse as they fold. The folding of an extended amino acid chain into a compact protein occurs because the final folded form is more energetically stable. But as part of this, each intermediate form seen along the way will usually be closer to the native state than its immediate predecessor. This downward progression is a funnel in a multidimensional space because there is more than just one way down, i.e., to get to the final form X, the amino acid chain might go through intermediates A, B, C or P, M, Q or S, T, U etc, or some hybrid mix of such routes. This approach has led to us understanding many cases of protein folding.
Is there any one paper or series of papers that led to this breakthrough?
In 1987, there was the first article by Bryngelson and Wolynes that set out the landscape picture. Later work by Onuchic and collaborators found the funnel shape of this landscape.
And everyone agrees with this perspective now?
More or less, yes. Earlier, everyone had been worrying about which specific pathway or route a protein would take to fold or unfold. The energy landscape theory said that each protein was likely to solve the search problem in its own way, but in several simple proteins you could calculate what the solution to this search problem was.
What findings led to this breakthrough?
The person who initially put forth this approach, Peter Wolynes, is an expert on glasses and he and Bryngelson used concepts from spin glass formation to understand protein folding. Two pieces of glasses may look the same to us, but microscopically they are very different: molten glass does not necessarily solidify by the same route every time, nor does it achieve its most stable state. It usually get "stuck " in one or another of an assortment of what we could call "stable enough" states, or local minima in the terminology of energy landscapes. A glass blower can push for more or less stability by cooling (quenching) the heated glass at different rates.
The various intermediate states of protein folding were that protein's own assortment of "stable-enough" states. The big difference however was that proteins only get stuck in these intermediate states temporarily: they have to converge on a single, functional form which lies at the bottom of the funnel. The specific sequence of the protein ensures that not too many deep minima exist along the way and thus there is enough momentum, as it were, for the folding protein to deal with the various troughs and bumps it will encounter on the funnel's surface. A protein will have acquired an amino acid sequence during evolution that allows this to occur, any random sequence will not fold.
Some of your own work has focused on the biological significance of those bumps and troughs in the funnel.
Yes, one might wonder why evolution has not led to these features which frustrate (and slow down) folding disappearing altogether. It is really an optimisation problem: for any given protein there will be a trade-off between ease of folding and functionality. Functionality will always be the top priority, while slower folding speeds may be fine for most cellular processes.
This line of thinking leads to the hypothesis that those parts of a protein that fold particularly slowly probably have some functionality and puts an extra tool at our disposal for identifying functional regions.
Are there any situations where faster folding is more important?
Yes, one of the dangers of slower folding is that partially folded proteins have a greater tendency to aggregate. Aggregation of proteins in the cell can not only lead to loss of function but also to protein misfolding diseases.
After six years at the University of California San Diego, you came here. What led to that and how has it been at NCBS?
I was applying across the board for positions, but India seemed more attractive than the US, where funding was going down. It's been good at NCBS. I really appreciate having a Faculty position, independence is a good thing.
The transition to NCBS has been very smooth. I need a lot of computing power. While I was getting ready to order my own cluster, Prof. Upi Bhalla was kind enough to give me access to his cluster. That helped a lot.
Your CV mentions that you have built up your own clusters in the past, and have programming skills. Are these necessary capabilities in the field?
I haven't really built a cluster, just administered a bunch of machines. That experience does help when things go wrong.
And how many people are working with you now?
I have three PhD students, one JRF and an intern. Oh and I do have one experimentalist in the group, that's my excursion into new territory.
I noticed on your website you say: "I don't do experiments".
True, but I don't avoid interacting with people who do! Also, there is the student who does experiments out of Prof. Jayant Udgaonkar's lab. That's been really good because I don't have the expertise to solve problems with experiments. We are working on a hyperthermophilic protein that has evolved to fold between 110-120 deg C. It's been an interesting protein and the project will likely turn out to be completely different from what we originally planned.
Have you had any Eureka moments in your scientific career?
No, I haven't. But I have had results that have slowly sunk in and been special. At first it felt like things had not worked but then I'd come around to seeing them in a different light. That's happened both during my PhD and postdoctoral work.
Here's a hypothetical question: It's many decades in the future, and you are about to retire. Is there anything that would make you look back and say: What an amazing life!
That is a hard question. I think it's really the luck of the draw, meaning I hope to be lucky enough to find and solve a problem that makes a big impact, but whether it will happen is another thing. More likely, one will make lots of small contributions but whether this will be satisfying as having, say, one big hit, who knows. Likely will be.
Any advice for people who are thinking of coming into science?
I would say just start thinking about problems. What I see with a lot of students, is that they come in wanting to be led somewhere. They want to be told what to do next, what experiment or calculation is needed. But it gets more interesting when it's your thought that is driving the project. It becomes your problem rather than someone else's and gives you a sense of achievement. I understood this thanks to my PhD supervisor: he was aware of what I was doing, but he would let me be. And it was a nice way of doing things. Students should start thinking about problems that they like. Sure, some of your ideas might be a bit crazy. On the other hand they might just need fine-tuning. The real enjoyment comes from solving a problem by yourself.
Comments
Post new comment